
Share on email
Share on linkedin
Share on twitter
Share on skype
Share on facebook
By Martin Willer
January 8, 2023
By Martin Willer
January 8, 2023
Since interest in yeasts began in the late 18th century, the lifecycle of a few species has been studied intensely. Many yeast cells have a well-established system for synthesis and secretion of small peptides. That’s why when DNA technology reached a point in the late 1980’s when heterologous expression became possible, one of the first choices of secretion vehicle was the alpha-factor from Saccharomyces cerevisiae.
Due to the very successful application of alpha-factor signals, it quickly became the expression system of choice within yeast labs. And expression systems using the alpha-factor setup still dominate heterologous expressions in yeast today.
Importantly, alpha-factor is almost exclusively translocated by the post-translational pathway.
However, there is another frequently overlooked translocation pathway that may be a better fit for your project: co-translational translocation. Keep reading to learn more about both alternatives and what you should consider when choosing the best translocation pathway.
Protein biosynthesis in living cells starts in the nucleus. Stretches of DNA (genes) are transcribed into RNA by enzymes called RNA polymerases and exported to the cytosol. Next, large RNA-protein complexes called ribosomes translate the RNA into proteins. For those proteins destined for secretion, translocation across a lipid bilayer (membrane) into the endoplasmic reticulum (ER) is the first step on the secretory pathway which ultimately leads to secretion of the protein to the exterior of the cell.
In yeast there are two functionally distinct pathways into the ER: the co-translational translocation pathway and post-translational translocation pathway.
The first pathway is the co-translational translocation pathway (Figure 1). The prefix “co” indicates that translocation takes place SIMULTANEOUSLY with translation. The ribosomes sit on the cytosolic side of the ER membrane and translate the RNA into protein while the protein is being translocated across the membrane into the ER lumen.
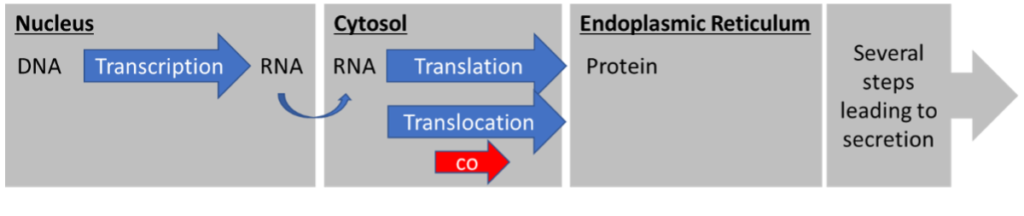
The second pathway is the post-translational translocation pathway (Figure 2). “Post” specifies that translocation happens AFTER translation. The entire protein is translated in the cytosol and only then translocated across the membrane, into the ER.
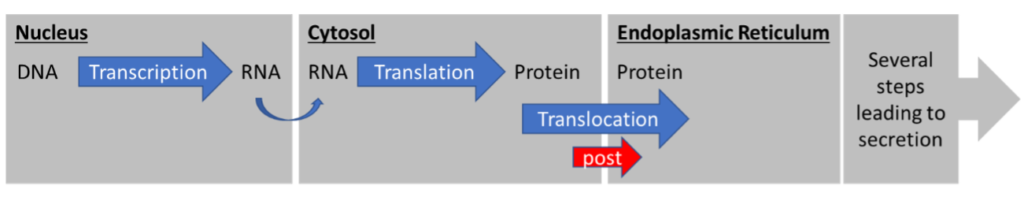
Two major differences between these pathways are instantly obvious. First, the protein is never exposed to the cytosol in co-translational translocation. Second, the transfer mechanisms across the ER membrane are different.
These differences should be accounted for when choosing the best translocation pathway for heterologous protein expression in yeast.
Pharma IT’s CMC team are ready to guide you through the process of choosing the right expression system for your project, ensuring a robust Master Cell bank! Our upstream consultants have decades of experience with Heterologous Protein Expression. At the same time our consultants can help you within QA and QC.
Pharma IT’s CMC team are ready to guide you through the process of choosing the right expression system for your project, ensuring a robust Master Cell bank! Our upstream consultants have decades of experience with Heterologous Protein Expression. At the same time our consultants can help you within QA and QC.
For example, alpha-factor is almost exclusively translocated by the post-translational pathway. Because alpha-factor is very small and highly soluble, this works well. However, suppose you were trying to express a larger, complex molecule with bulky folding and perhaps hydrophobic domains, and then having to translocate it across a lipid membrane from the cytosol into the ER after synthesis? It does not require much imagination to predict potential problems.
For this reason, it is important to remember that co-translational translocation is an alternative option.
It is also important to mention that in many cases it is possible to redirect the protein translocation from one pathway to the other.
At Pharma IT, we can guide you through these considerations and help you ensure a robust Master Cell bank.
A combination of predictive tools and experimental data will form the basis for our recommendations of the optimal pathway choice. Our team has experience with these types of analysis as well as mitigating actions. You are always welcome to contact our team to discuss concrete cases.
Martin Willer is a Senior Pharma Consultant who holds over 30 years of experience within molecular biology, cell biology, and biotechnology in both academic and industrial settings. His project involvement has generally fallen into two main categories: heterologous expression and metabolic pathway engineering. Martin has worked extensively with different yeast platforms and has experience with most pro- and eukaryotic systems. He holds a PhD in Biology and Genetics from the University of Copenhagen.